Project: Development of the MEDomicsLab platform
Status
In progress (2020-today)
Type
General
Team
- Andréanne Allaire1 (2022-2024)
- Brahim Fakri1 (2022-2025)
- Cedrik Lampron1 (2022-2025)
- Charles Lévesque-Matte1 (fall 2020)
- Charles-Olivier Ipperciel1 (2024-today)
- Clarisse Cheng1 (summer 2022)
- Corentin Gauthier1 (summer 2022)
- Guillaume Blain1 (summer 2022-today)
- Hithem Lamri1 (fall 2022-2023)
- Jonathan Perron1 (2020-2021)
- Kayla Davio-Roy1 (summer 2021)
- Ludmila Amriou1 (2023-2024)
- Lyna Hiba Chikouche1 (2023-2024)
- Mahdi Ait Lhaj Loutfi1 (2021-today)
- Mamadou Mountagha Bah1 (2020-2021)
- Mariem Kallel1 (2024-today)
- Mohammed Benabbassi1 (fall 2022)
- Nicolas Longchamps1 (summer 2022-today)
- Ouael Nedjem Eddine SAHBI1 (2023-today)
- Robin Mailhot1 (fall 2020)
- Sarah Denis1 (2023-2024)
- Martin Vallières1 (2020-today)
1 Department of Computer Science, Université de Sherbrooke, Sherbrooke (QC), Canada
Description
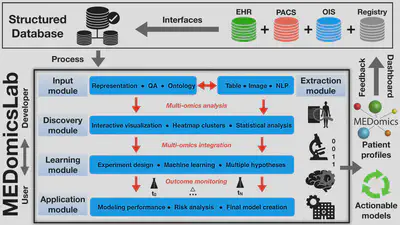
MEDomicsLab is an open-source computing platform for integrative data modeling in medicine. It was created by an international consortium of medical scientists (https://medomics.ai), whose main goal is to facilitate the development and clinical translation of artificial intelligence (AI) applications in medicine. The platform is based on the Python programming language and contains five major modules that allow loading, processing, exploring multi-omics data, and creating and evaluating actionable models for precision medicine. MEDomicsLab’s intrinsic workflow is designed to provide different levels of abstraction of methodological complexity to users and developers via application scripts, option parameters and class structures. In general, MEDomicsLab is envisioned to be at the center of multidisciplinary research teams and hospital database infrastructures.
Recent advances in so-called “omics” technologies (genomics, radiomics, proteomics, etc.) offer unprecedented opportunities to characterize biological processes that are related to certain phenotypes. The effective combination of these elements as “multi-omics modeling” will certainly allow us to better tailor treatments to individual patients (i.e., “precision medicine”).
Given the complexity of medical problems, achieving the full potential of precision medicine is directly related to our ability to properly manage, structure and leverage our hospital databases. Therefore, mastering and combining the following data analysis techniques should increase the impact of the “multi-omics” prediction models that are built from these databases:
- Image analysis: extraction of radiomic features allows us to better characterize tumor heterogeneity. The MEDimage package is the dedicated component for this task.
- Machine learning: some learning techniques allow us to better combine data from different categories (such as multi-omics).
- Deep learning: the multitude of deep neural network architectures offers several possibilities for the automatic learning of different tasks in medicine. The performance of some networks is sometimes superior to that of humans.
- Automatic text processing: automatic processing and coding of textual reports from electronic patient records would allow for better integration of information relevant to multi-omics modeling throughout the patient management period.
- Federated learning: in order to increase the amount of modeled data from different health care institutions, a distributed learning approach can be used - data is kept within the boundaries of each institution, thus avoiding data transfer and privacy issues.
For more information, see the project website.
This platform is being developed in collaboration with:
- University California San Francisco (Olivier Morin, Taman Upadhaya, Jorge Barrios)
- University of Toronto (Jan Seuntjens)
- The D-Lab (Philippe Lambin, Henry Woodruff, Avishek Chatterjee)
- Oncoray (Alex Zwanenburg, Steffen Löck)
- Université Laval (Louis Archambault, Philippe Després)